AG Becker
Our research group studies the cellular reaction of algal cells to the environment. Major research topic is the regulation of water homeostasis in various systems. Using classical cell biological (video microscopy, fluorescence microscopy, protein biochemistry) and forward and reverse genetic approaches we are investigating the role of the contractile vacuole in osmoregulation in Chlamydomonas. Furthermore, using omics approaches we investigate desiccation tolerance but also the effect of light and temperature stress in lab experiments and natural alpine and polar habitats
AG Bucher
Mechanisms of Mineral Nutrient Acquisition in Plants
The Bucher Lab studies the molecular basis of symbiotic plant-microbe interactions. Most of our work focuses on the arbuscular mycorrhizal symbiosis (AMS) which is based on an intimate interaction between most vascular plants and soil fungi from the phylum Glomeromycota.

AG de Meaux
Adaptive molecular variation in plant systems
Our research seeks to reconstruct the recent history of adaptive molecular variation in plant systems. In other terms, we aim at dissecting the molecular mechanisms of Darwinian evolution in complex natural systems. We work with the weedy annual and model plant species Arabidopsis thaliana and its close relatives A. lyrata and A. halleri. Our work focuses on understanding links between life history strategies and fitness. We further intend to develop new methods to track the action of natural selection at the molecular level.
AG Döhlemann
Molecular mechanisms of microbe - plant interactions
Our research aims to identify and understand molecular mechanisms of microbe-plant interactions. Focus is on effector proteins of biotrophic microbes, which suppress host immunity and plant metabolism.
Within CEPLAS, we are interested to i) study the role of plant cysteine proteases in immunity and how they are modulated by microbial effectors and ii)elucidate how organ-specific factors of both plant and colonizing microbe contribute to interaction outcomes.
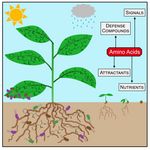
AG Hildebrand
Amino acid metabolism in plant development and environmental interactions
Amino acids are involved in several physiological processes apart from constituting proteins. Plant amino acid metabolism strongly reacts to challenges caused by both, biotic as well as abiotic stress conditions and provides a diverse set of signaling and defense compounds. Using a combination of biochemical and omics approaches we aim to identify presently unknown reaction steps in amino acid metabolism as well as the biochemical mechanisms of amino acid sensing and signaling.

AG Höcker
Protein Degradation in Light-controlled Plant Development
We are studying the role of protein degradation in light-controlled plant development. To unravel this process, we are using the model species Arabidopsis thaliana and a combination of genetic, molecular and biochemical methods.
AG Hülskamp
Molecular Cell Biology and Developmental Genetics
We use Arabidopsis trichomes as a model system to study cell-cell communication, cell differentiation and morphogenesis. Genetic screens have revealed a large number of mutants affecting distinct steps of trichome development enabling a further molecular and cell biological analysis. The beauty of this system is that virtually all trichome genes turned out to be relevant for other cell types as well because they are involved in various general mechanisms of plant development
AG Kopriva
Plant mineral nutrition
The long term goal of our research is to understand how plants integrate the uptake and utilization of key mineral nutrients with their needs, demand, and changes in environment. We use a combination of biochemical, genetic and physiological approaches and exploit natural variation in the model species Arabidopsis thaliana. We defined the transcriptional mechanisms controlling sulfate uptake and assimilation and showed how this is linked to uptake of other nutrients such as nitrate (reviewed in Takahashi et al., 2011).
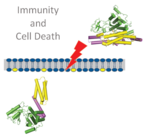
AG Maekawa
Immunity and cell-death
Over the past decades, growing evidence has highlighted remarkable similarities between the innate immune systems of the plant and animal kingdoms. These ‘shared’ innate immune systems include intracellular receptors as exemplified by nucleotide-binding leucine-rich repeat proteins (NLRs). Our group aims to unravel the underlying mechanistic parallels between plant and animal immune components using plants as a model. Our research programme encompasses a diverse array of experimental systems including genetics, genomics and cellular and structural biology with an emphasis on immunity and cell death.

AG Saur
Plant-microbe interactions
Our research focuses on the molecular mechanisms underlying the interaction of pathogenic microbes with their plant hosts. We particularly focus on understanding how molecules (so-called AVRa effectors) from the powdery fungus facilitate successful infection of the barley host and the development of fungal diseases on cereals. Our research aims to contribute to the control fungal phytopathogens on economically important crops.
AG Stetter
Crop evolution
Our research group is interested in plant adaptation and evolution. We use the domestication of crops as model to study how plants respond to changing environments. Our models are, the incomplete domestication of the South American pseudo-cereal amaranth, and the domestication and cultivation history of maize. We use population and quantitative genetic methods to understand how wild plants became crops and how these crops spread across the globe.

AG Thomma
Evolutionary microbiology: fungal plant pathogens
Our research aims to identify mechanisms that underly pathogenicity of fungi on plant hosts. To this end, we study the soil-borne broad host-range vascular wilt fungus Verticillium dahliae and try to understand molecular processes that mediate adaptation to plant hosts by studying the evolution of “effector activities” that are exerted by fungal secreted molecules to mediate host colonization. The functional analysis of the most relevant effector proteins leads to the discovery of crucial processes that are targeted by the fungus to subvert host immunity and support host colonization. One of the most recent discoveries concerns the identification of Verticillium dahliae effector proteins secreted during host colonization that manipulate the host microbiome to particularly repress microbial antagonists.
AG Töpfer
Plant Metabolic Systems and Their Interactions
Our research aims to gain a better understanding of the behaviour of plant metabolic systems and their interactions. Our group uses computational approaches that are centred around the analysis of large-scale metabolic networks and works closely with experimental labs. Key topics are the development of flux-balance methods to study tissue- and organ interactions, the curation and computational integration of specialised metabolism and the study of plant-environment interactions on a biotic and abiotic level.

AG Zuccaro
Functional genomics and molecular biology of symbiotic fungi
Research in our group focus on the mechanisms that enable symbiotic fungi to colonize plants successfully and on the processes accounting for variations in host preferences and fungal lifestyles. The prime models for our studies are the root endophyte Piriformospora indica (Basidiomycota, Sebacinales), and the orchid mycorrhizal Sebacina vermifera, two biotrophic symbiont that colonizes the root epidermal and cortex cells of a broad range of plant species, including the dicot model plant Arabidopsis thaliana and the agriculturally important monocot Hordeum vulgare.